Abstract
Understanding factors that shape the immune landscape across hematological malignancies is essential for immunotherapy development. How cancer-cell intrinsic genomic and epigenetic alterations influence immune signatures in hematological malignancies is not known. Here, we integrated over 8,000 transcriptomes of hematologic cancers and multilevel genomic datasets to investigate associations of immune states to cancer molecular subtypes, genetic and epigenetic alterations, and clinical outcomes.
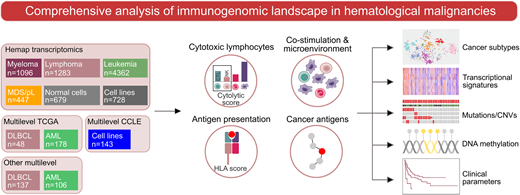
We utilized a resource of over 8,000 transcriptomes collected across 36 hematologic malignancies and normal hematopoietic cells (Hemap), together with multi-omics datasets of acute myeloid leukemia (AML) and diffuse large B-cell lymphoma (DLBCL) from The Cancer Genome Atlas and other sources (Figure). In addition to gene expression data, we integrated somatic DNA alterations, methylation data, multiplex immunohistochemistry (mIHC), and flow cytometry to comprehensively map immune-associated features and validate the robustness of the findings. To characterize the composition of the cytolytic immune infiltrate from bulk transcriptomes, we defined a signature of genes most specifically expressed in cytotoxic CD8+ T lymphocytes and natural killer (NK) cells termed cytolytic score.
We found significant heterogeneity in the cytotoxic lymphocyte infiltration signature across hematologic malignancies. Highest cytolytic infiltrate was detected in lymphomas and correlated with IFN-γ and myeloid cell infiltration signatures including CXCL9-11 and IDO1, distinguishing the lymphoma microenvironment from leukemias. In addition to transcriptomic microenvironmental properties, specific genetic alterations were associated with cytotoxic lymphocyte infiltration. In DLBCL, driver alterations enriched in the germinal center B-cell like (GCB) molecular subtype including BCL2 translocations and KMT2D were linked to an immune-cold transcriptomic phenotype. In contrast, DTX1 alterations defined immune-infiltrated lymphomas within the GCB molecular subtype. In AML, TP53 mutations and complex karyotype were enriched in a distinct tSNE-based transcriptomic cluster characterized by increased immune infiltration in the bone marrow (BM).
Given the importance of effective antigen presentation for adaptive anti-tumor immune responses, we aimed to understand the transcriptional regulation of HLA genes and co-stimulatory and co-inhibitory signaling in subtypes of hematological malignancies. Downregulation of the antigen-presenting HLA II genes was associated with CpG methylation of the promoter region of the HLA class II master regulator CIITA in distinct transcriptomic clusters of AML harboring PML-RARA or NPM1 alterations. Expression of genes encoding immune checkpoint molecules was strongly influenced by the cell-of-origin and microenvironment of each cancer type. We identified novel associations of inhibitory immune checkpoint molecules to disease subtypes, such as VISTA/PD1-H enriched in myeloid malignancies including AML, CML, and MDS, validated by mIHC performed on BM biopsies. Furthermore, variation in the expression of several genes encoding immune checkpoints was associated with somatic mutations (e.g. CD70 in DLBCL), copy-number alterations (e.g. MICB in DLBCL), and DNA methylation (e.g. PDL1 and PDCD1LG2 in AML).
Finally, we integrated GTEx gene expression data across tissues to define cancer-germline antigens (CGAs) with an immune privileged tissue expression pattern. CGAs were frequently expressed in multiple myeloma and DLBCL compared to other hematologic malignancies. CGA expression was associated with cytogenetic alterations and increased MYC activity signature in myeloma and CD58 and KLHL6 mutations in DLBCL. In addition, CGA expression in myeloma and DLBCL was linked to reduced antigen gene promoter methylation and decreased survival.
In summary, our findings demonstrate that molecular subtypes of hematological malignancies harbor distinct immunological signatures influenced by genetic and epigenetic alterations. Integrating genetic, epigenetic, and transcriptomic data may facilitate the development of precision immune intervention strategies in hematological malignancies.
Leppa:Bayer: Research Funding; Celgene: Consultancy; Roche: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Research Funding; Takeda: Consultancy, Research Funding. Mustjoki:Bristol-Myers Squibb: Honoraria, Research Funding; Celgene: Honoraria; Pfizer: Honoraria, Research Funding; Ariad: Research Funding; Novartis: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal